Diversity | Free Full-Text | SSR Markers for Trichoderma virens: Their Evaluation and Application to Identify and Quantify Root-Endophytic Strains
Development of novel EST-SSR markers for ploidy identification based on de novo transcriptome assembly for Misgurnus anguillicaudatus | PLOS ONE

General flowchart of SSR marker discovery using different sequencing... | Download Scientific Diagram
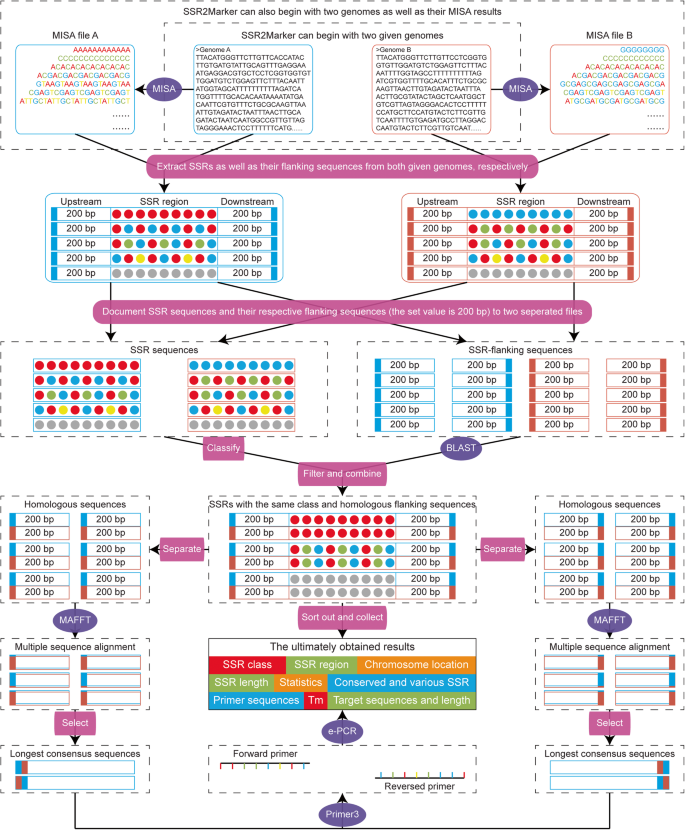
SSR2Marker: an integrated pipeline for identification of SSR markers within any two given genome-scale sequences | Molecular Horticulture | Full Text

Comprehensive functional analysis and mapping of SSR markers in the chickpea genome (Cicer arietinum L.) - ScienceDirect
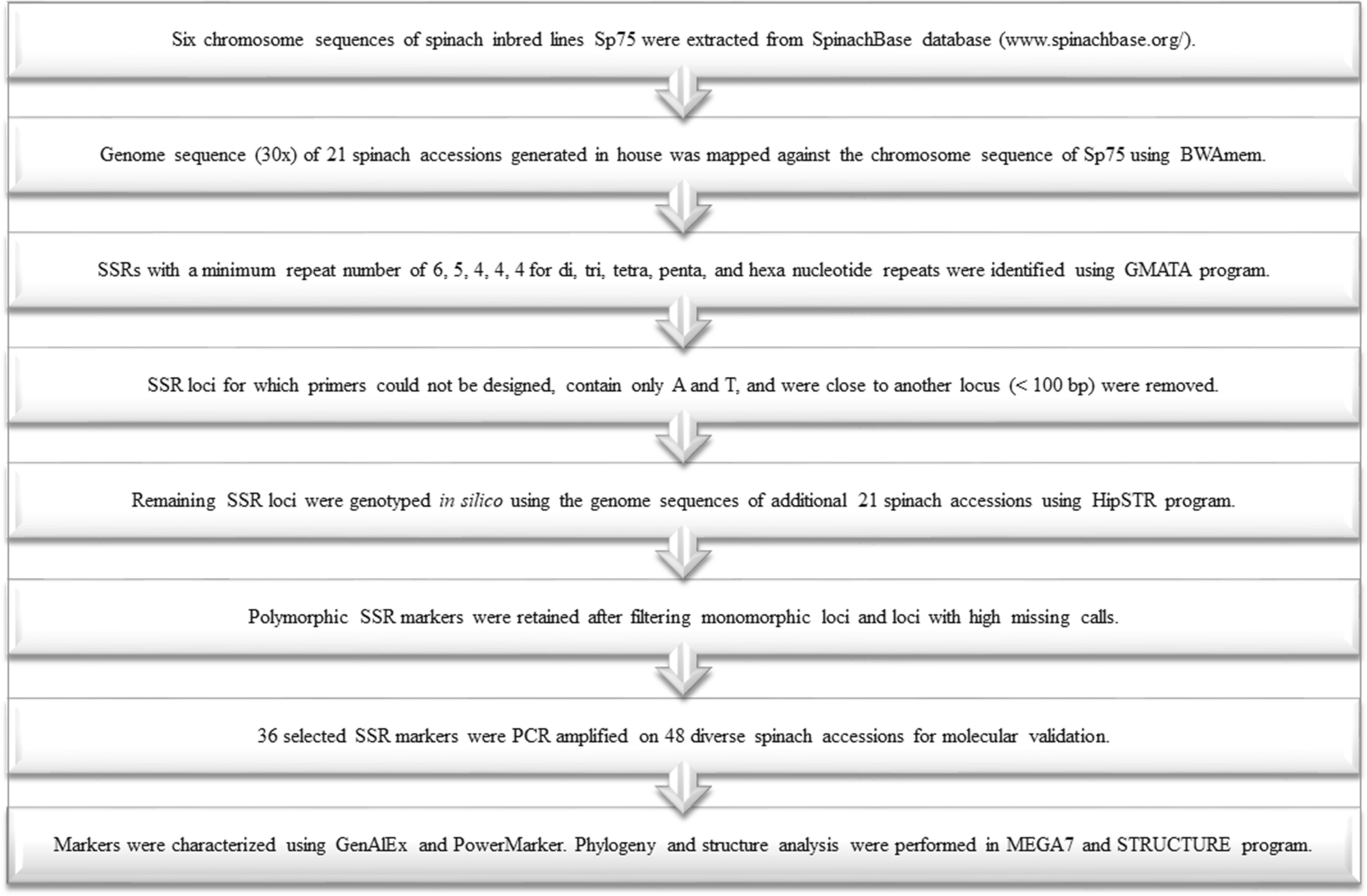
Genome-wide simple sequence repeats (SSR) markers discovered from whole-genome sequence comparisons of multiple spinach accessions | Scientific Reports

A general protocol for developing SSR markers with a SSR-enrichment step. | Download Scientific Diagram
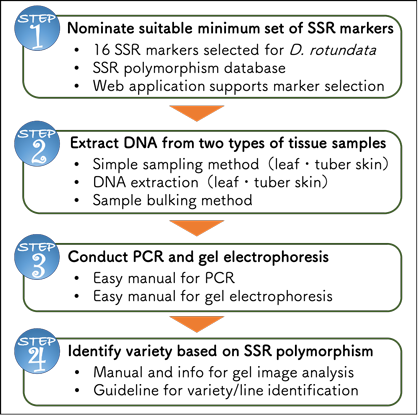
SSR marker technology package for variety identification of white Guinea Yam | Japan International Research Center for Agricultural Sciences | JIRCAS

Characterization and Development of EST-SSR Markers from Transcriptome Sequences of Chrysanthemum (Chrysanthemum ×morifolium Ramat.) in: HortScience Volume 54 Issue 5 (2019)

A general protocol for developing SSR markers with a SSR-enrichment step. | Download Scientific Diagram
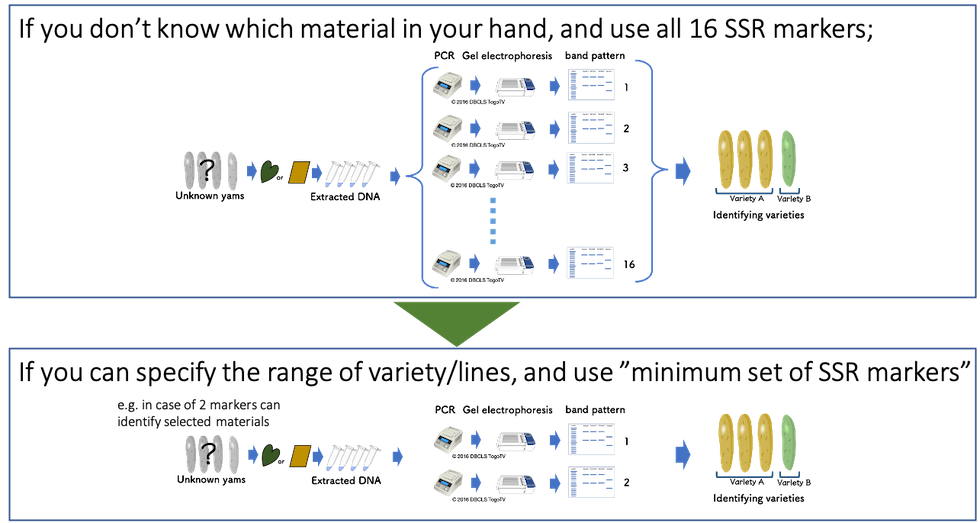
Yam Variety Identification Toolkit:Step 1 | Japan International Research Center for Agricultural Sciences | JIRCAS

EST-SSR markers derived from an elite barley cultivar (Hordeum vulgare L. 'Morex'): polymorphism and genetic marker potential. | Semantic Scholar
![The newly developed genomic-SSR markers uncover the genetic characteristics and relationships of olive accessions [PeerJ] The newly developed genomic-SSR markers uncover the genetic characteristics and relationships of olive accessions [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2020/8573/1/fig-2-1x.jpg)
The newly developed genomic-SSR markers uncover the genetic characteristics and relationships of olive accessions [PeerJ]
![Frontiers | New Hypervariable SSR Markers for Diversity Analysis, Hybrid Purity Testing and Trait Mapping in Pigeonpea [Cajanus cajan (L.) Millspaugh] Frontiers | New Hypervariable SSR Markers for Diversity Analysis, Hybrid Purity Testing and Trait Mapping in Pigeonpea [Cajanus cajan (L.) Millspaugh]](https://www.frontiersin.org/files/Articles/243873/fpls-08-00377-HTML-r1/image_m/fpls-08-00377-g001.jpg)
Frontiers | New Hypervariable SSR Markers for Diversity Analysis, Hybrid Purity Testing and Trait Mapping in Pigeonpea [Cajanus cajan (L.) Millspaugh]
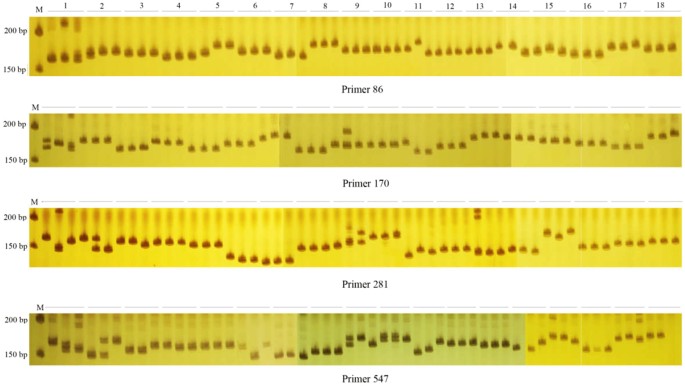
Cross-species transferability of EST-SSR markers developed from the transcriptome of Melilotus and their application to population genetics research | Scientific Reports

Development of Genome-wide SSR Markers for Physical Map Construction with PCR-based Polymorphic SSRs in Jute (Corchorus Spp.) | SpringerLink